Evolution of gene expression in Siphonophores
An important challenge in comparative biology is comparing functional genomic data across species. Comparisons of gene expression are critical for understanding genotypic and phenotypic evolution, and also for investigating the extent to which expression data obtained in model organisms can be extrapolated to related species. There are several major challenges to these comparisons: 1) Which expression metric should we compare across species? 2) How do we normalize expression data across both tissues and species? 3) How do we address gene homology and phylogenetic relationships in order to make meaningful comparisons of genes and gene expression across species?
Using gene expression data collected from 5 zooids and 1 tissue, I investigated gene expression patterns in homologous zooids/tissues across 7 siphonophore species, and present three possible solutions to the challenges outlined above. This work is available in Molecular Biology and Evolution. We first investigated expression patterns in a classical manner within species using standard differential gene expression methods. This enables the investigation of expression patterns among novel zooids that are specific to particular species, such as the tentacular palpon in Physalia physalis.
I then addressed the three challenges outlined above: 1) Proposed a metric to account for differences in sequencing depth across species. 2) Used ratios of expected counts to account for unknown species- and gene-specific counting-efficiency coefficients (see Dunn et al., 2013). 3) Proposed a new solution to the third challenge to comparisons of expression across species: a fully phylogenetic approach that we call Species Branch Filtering (SBF), where we map expression data to gene trees. I compared SBF with a ‘classical’ approach of comparing gene expression patterns found in strict orthologs. By contrast with SBF, this type of analysis focuses exclusively on genes that have very specific evolutionary histories, and discards a number of genes with more complex histories that may be of interest to the investigator. We call this classical approach Species Tree Filtering (STF).
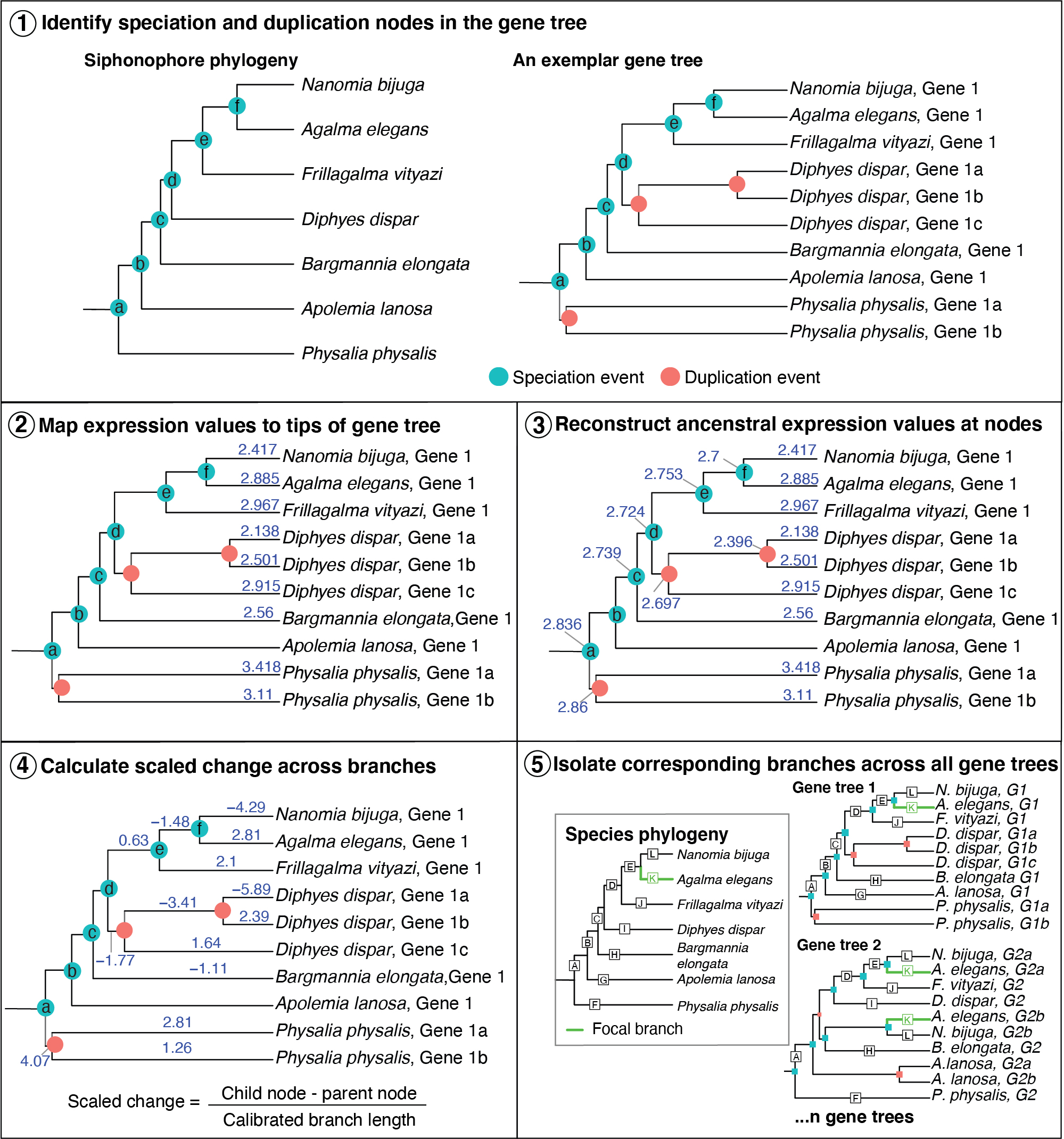
Figure: New phylogenetic approach to identify evolutionary changes in gene expression, called Species Branch Filtering (SBF). Step 1, we label each of the nodes in the species tree, and identify equivalent speciation nodes across every gene tree (an exemplar is shown here). Step 2, we map expression values (TPM10K) to the tips (expression values are mapped and reconstructed for each homologous zooid separately). Step 3, we reconstruct ancestral trait expression values at all internal nodes where expression data are available. Step 4, we calculate scaled change in gene expression (child node expression - parent node expression / branch length). Branch length is calibrated to the species tree branch lengths. Step 5, we identify branches in gene trees that correspond to equivalent branches in the species tree. There may be more than one branch in a gene tree that corresponds to the same branch in the species tree.
Using ‘classical’ strict ortholog methods we identified a number of zooid/tissue variable genes, as well as species variable genes. Using SBF, we find that the vast majority of species-equivalent branches show changes of expression close to 0 (neutral changes), suggesting that for closely related genes, expression patterns tend to be largely consistent across species. A subset of branches showed positive and negative changes in expression ratios across the branch, some very large, suggesting putative lineage-specific changes in expression.